PSS database schema
Naming conventions
- Name format
-
- Node labels: UpperCamelCase
- Relationship types: UPPER_SNAKE_CASE
- Property keys: snake_case
- Edge
- Relationship between two nodes
- Source
- source (start) node of a relationship.
- Target
- target (end) node of a relationship.
Node labels
A node can have multiple labels. In general, the primary labels have a constraint of a unique name within the label.
-
Primary Labels
Primary label Secondary labels (*=optional) Description Complex Node Complexes that can contain plant, foreign and/or metabolite components Metabolite Node, MetaboliteFamily* Chemical entities Family Node, [PlantAbstract | PlantCoding | PlantNonCoding] Gene family (set of all genes within the family) Clade Node, [PlantAbstract | PlantCoding | PlantNonCoding] Set of genes defined by sequence similarity FunctionalCluster Node, [PlantAbstract | PlantCoding | PlantNonCoding] Set of genes with the same function Process Node ForeignEntity Node, Foreign Biological foreign entity, such as a pathogen species ForeignCoding Node, Foreign Protein coding agent of an external factor, such as a pathogen gene ForeignNonCoding Node, Foreign Non-coding agent of an external factor, such as a pathogen gene ForeignAbstract Node, Foreign Undefined or unspecified agent of an external factor, such as a pathogen gene ForeignAbiotic Node, Foreign Abiotic factors (e.g. heat) Reaction Node Reaction node defines a reaction or interaction between entities Condition Condition that affects reaction(s) -
Secondary Labels
Label Description Node All nodes MetaboliteFamily A group of similar chemical entities (e.g. reactive oxygen species) PlantAbstract Undefined or unspecified plant gene (e.g. without standard gene identifier) PlantCoding Plant gene with protein gene product PlantNonCoding Plant gene without protein gene product (e.g miRNA) Plant All plant specific nodes (PlantAbstract, PlantCoding, PlantNonCoding) Foreign External factors that are neither plant nor metabolite related
Edge types
Each edge only has one type.
-
Hierarchical related edges
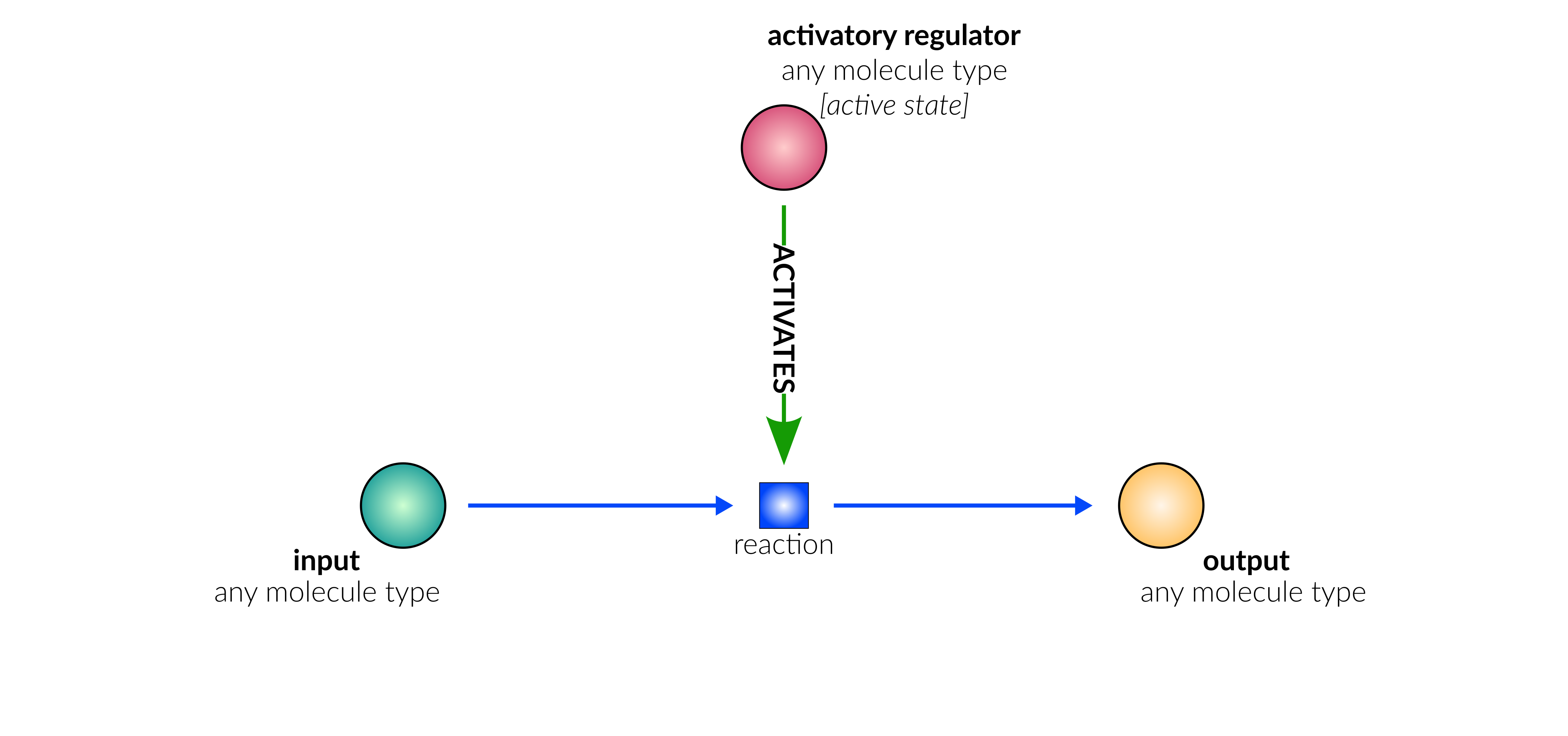
Type Description Edge example COMPONENT_OF Source node is a component of target node (Complex) (JAZ) ―[:COMPONENT_OF]→ (JAZ|DELLA) TYPE_OF Source node (Metabolite) is a type of target node (MetaboliteFamily) (O3) ―[:TYPE_OF]→ (ROS) AGENT_OF Source node (Foreign[*]) is an agent of target node (ForeignEntity) (VPg) ―[:AGENT_OF]→ (potyvirus) HAS_CLADE Source node (Family) has child clade target node (Clade) (AGO) ―[:HAS_CLADE]→ (AHP1,2,3,4,5) TAKES_PART Source node (Family) takes part in reactions via target node (FunctionalCluster) (ACS) ―[:TAKES_PART]→ (ACS6) CONDITION_INPUT Source node form part of the condition target node (Condition) defines (JAZ) ―[:CONDITION_INPUT]→ (NPR1 high & JAZ high) -
Reaction related edges
Type Description Edge example ACTIVATES Source node activates target node (Reaction) (CYP94) ―[:ACTIVATES]→ (rx00042) INHIBITS Source node inhibits target node (Reaction) (miR6022) ―[:INHIBITS]→ (rx00308) SUBSTRATE Source node is a substrate in the reaction, and is consumed/catalysed by reaction (JA-Ile) ―[:SUBSTRATE]→ (rx00042) PRODUCT Target node is a product in the reaction, and is produced/resulted from reaction (rx00042) ―[:PRODUCT]→ (12-OH-JA-Ile) TRANSLOCATE_FROM Source node is translocated from source_compartment by reaction (sister to SUBSTRATE) (SA) ―[:TRANSLOCATE_FROM]→ (rx00072) TRANSLOCATE_TO Target node is translocated to target_compartment by reaction (sister to PRODUCT) (rx00072) ―[:TRANSLOCATE_TO]→ (SA)
Properties
| Property name | Applies to (labels/edge types) | Type | Note |
|---|---|---|---|
| added_by | - | string | |
| creation_date | - | string | |
| name | - | string | |
| components | Complex | list | |
| classification | ForeignEntity | string | Phylogenetic classification |
| species | ForeignEntity | string | |
| family | FunctionalCluster | string | |
| sequence | FunctionalCluster | string | Only in case of gene not existing in genome model (of GoMapMan) |
| short_name | FunctionalCluster | string | |
| additional_information | Node | string | |
| curated | Node | bool | |
| external_links | Node | list | unknown, invented:<reason>, or <database>:<database identifier>, where database includes doi, pmid, aracyc, chebi, pubchem, kegg, etc |
| model_version | Node | string | |
| pathway | Node | string | One of defined pathways |
| model_status | Node excluding Reaction | string | |
| description | Node, Condition | string | |
| <species>_homologues | Plant | list | list of species specific homologues |
| species | Plant | list | Subset of defined species |
| evidence_sentence | Reaction | string | |
| experimental_techniques | Reaction | string | |
| reaction_effect | Reaction | string | activation or inhibition |
| reaction_mechanism | Reaction | string | |
| species | Reaction | list | Subset of defined species, and "all" |
| trust_level | Reaction | string | R1;R2;Rx... |
| reaction_id | Reaction, all reaction related edges | string | |
| reaction_type | Reaction, all reaction related edges | string | One of defined reaction types |
| source_form | ACTIVATES, INHIBITS, SUBSTRATE, TRANSLOCATE_FROM | string | One of defined node forms |
| source_location | ACTIVATES, INHIBITS, SUBSTRATE, TRANSLOCATE_FROM | string | One of defined locations |
| source_organ | ACTIVATES, INHIBITS, SUBSTRATE, TRANSLOCATE_FROM | string | One of defined plant organs (leaf, stem, root) |
| target_form | PRODUCT, TRANSLOCATE_TO | string | One of defined node forms |
| target_location | PRODUCT, TRANSLOCATE_TO | string | One of defined locations |
| target_organ | PRODUCT, TRANSLOCATE_TO | string | One of defined plant organs (leaf, stem, root) |
Indices
Indices are generated across nodes with the same label to allow faster searching.
In progress
Plant node hierarchy
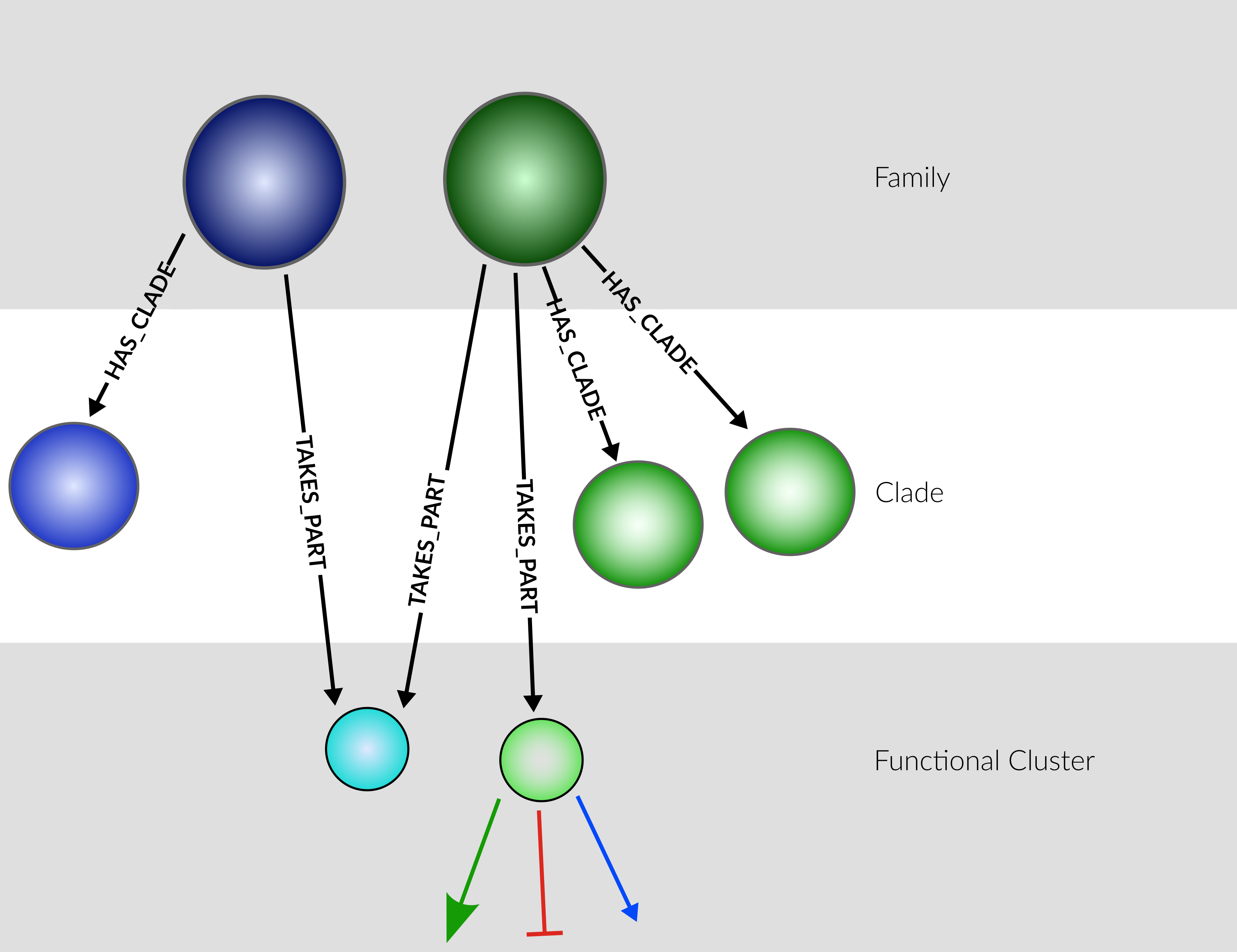
Families contain all genes associated with the family, across plant species.
Clades are subsets of families, as defined by sequence similarity.
FunctionalCllusters are subsets of (possibly multiple) families, as defined by participation in reactions (i.e. their function).
Of the three layers, only FunctionalClusters can participate in reactions.
Plans - incorporate Clades as "parents" of FunctionalClusters, instead of families
Defined reaction types
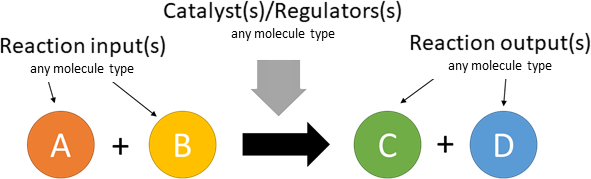
The database schema is based on a representations of chemical reactions. In a simple example, the following reaction A + B → C (catalysed by e):
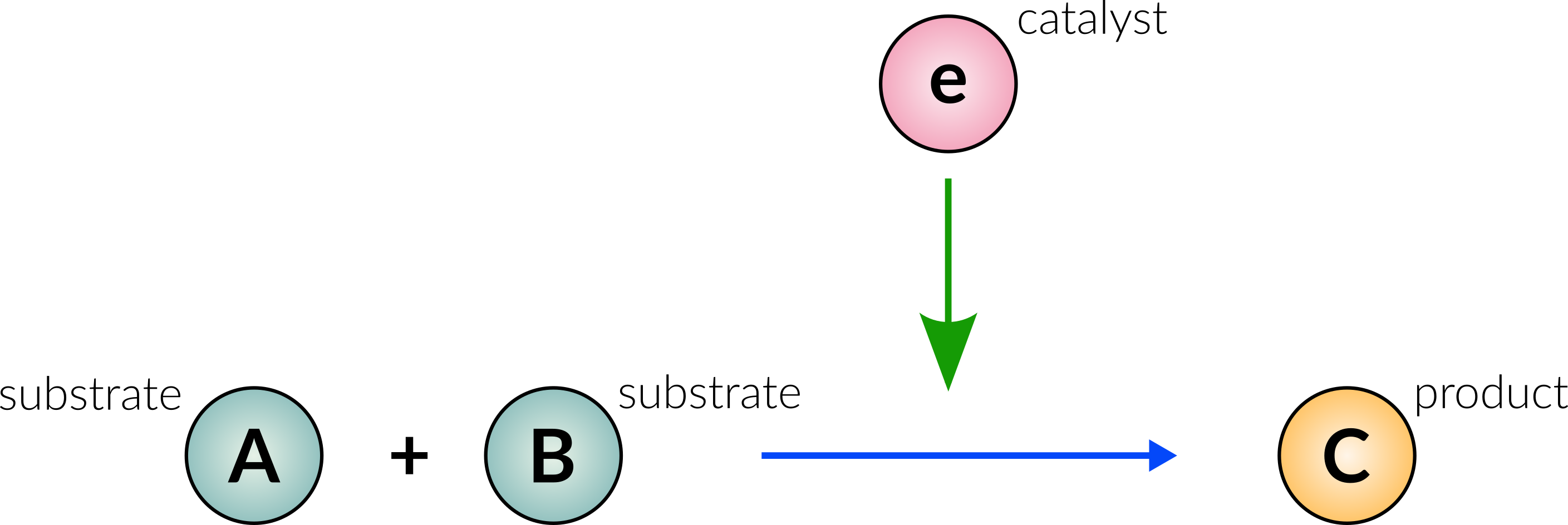
can be represented in the graph database (using only nodes and edges) as follows:
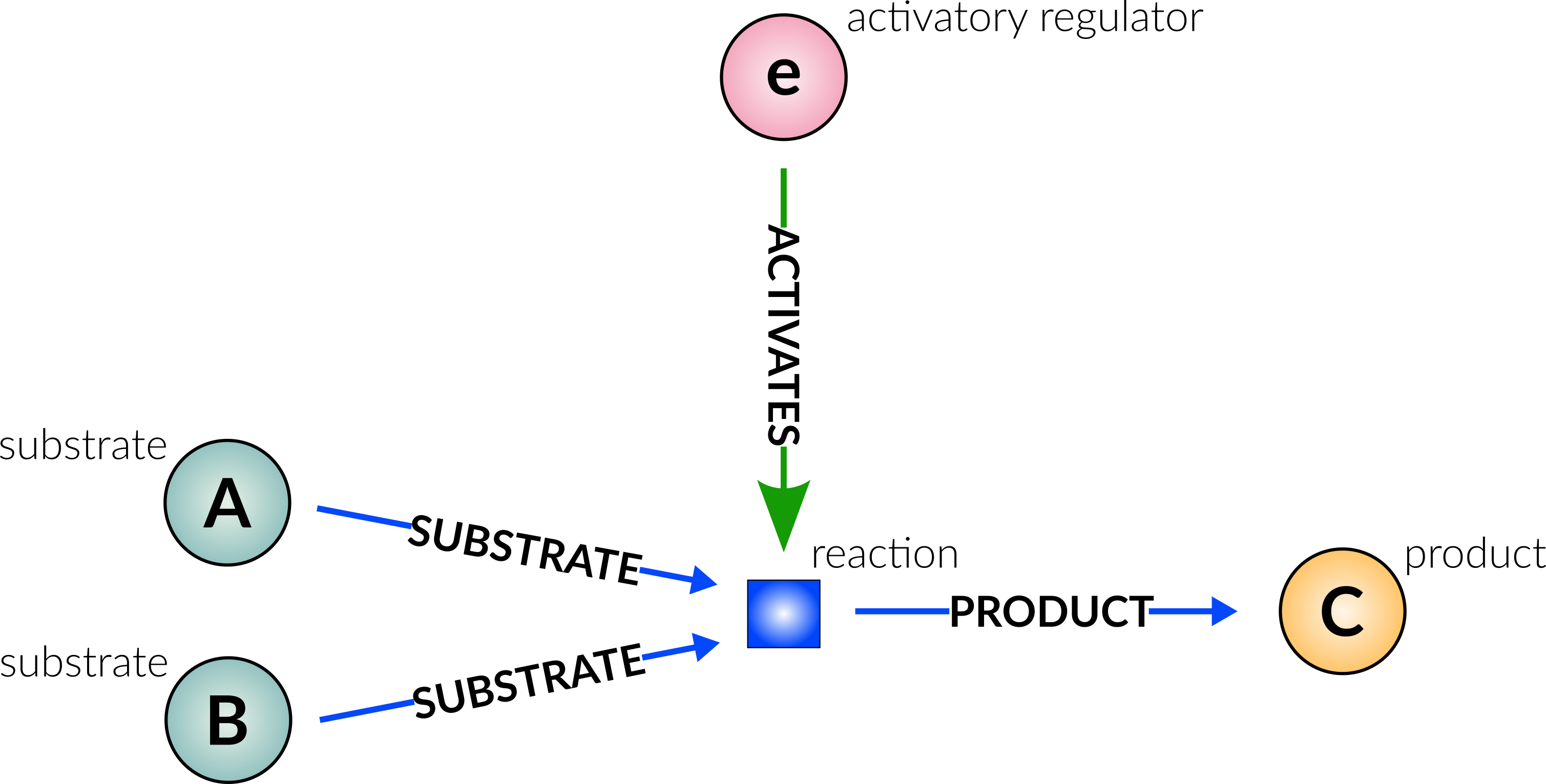
Edges (arrows) and reaction nodes are reaction specific. Nodes other than reactions can take part in multiple reactions.
Using this schematic, the database can contain the following reaction types:
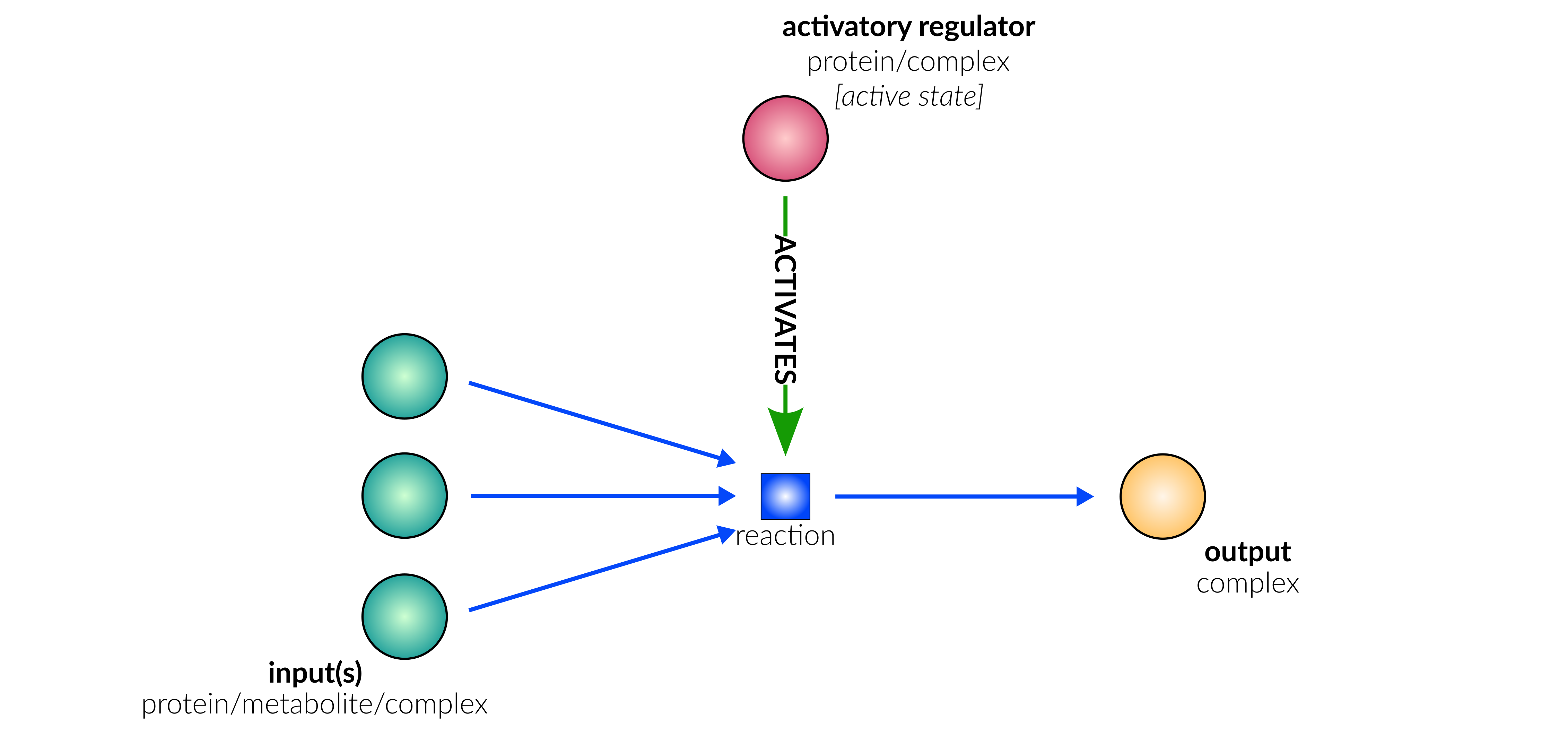
binding/oligomerisation
Biological representation
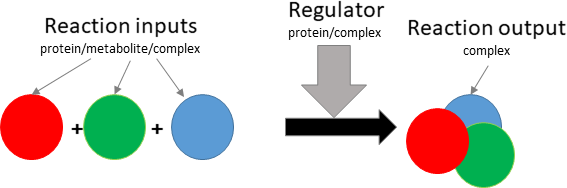
Database representation
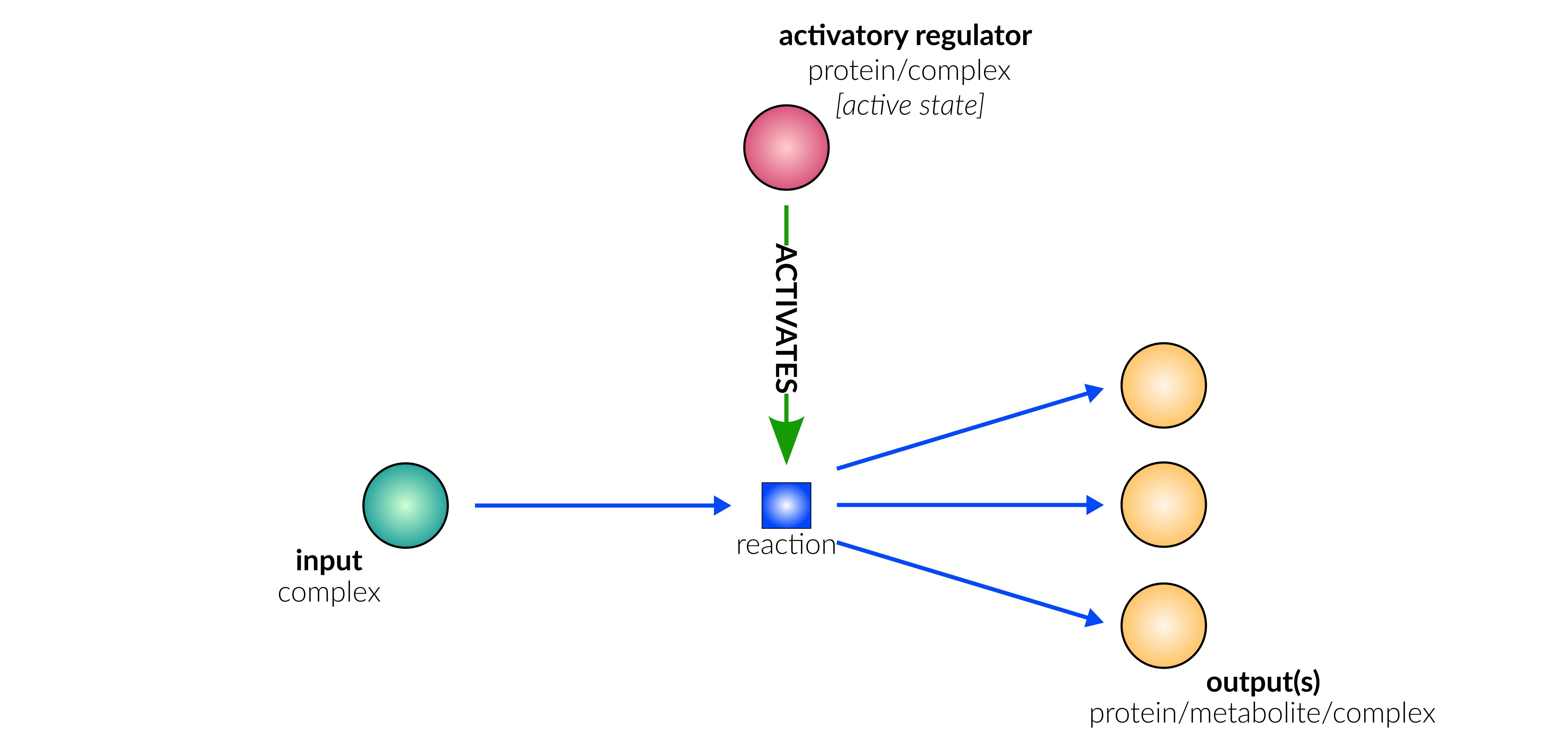
dissociation
Biological representation
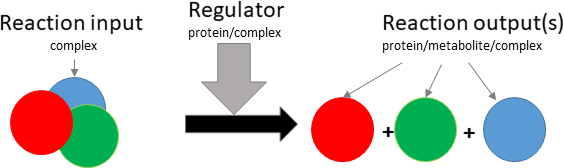
Database representation
catalysis
Biological representation
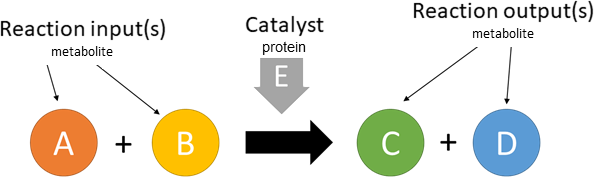
Database representation
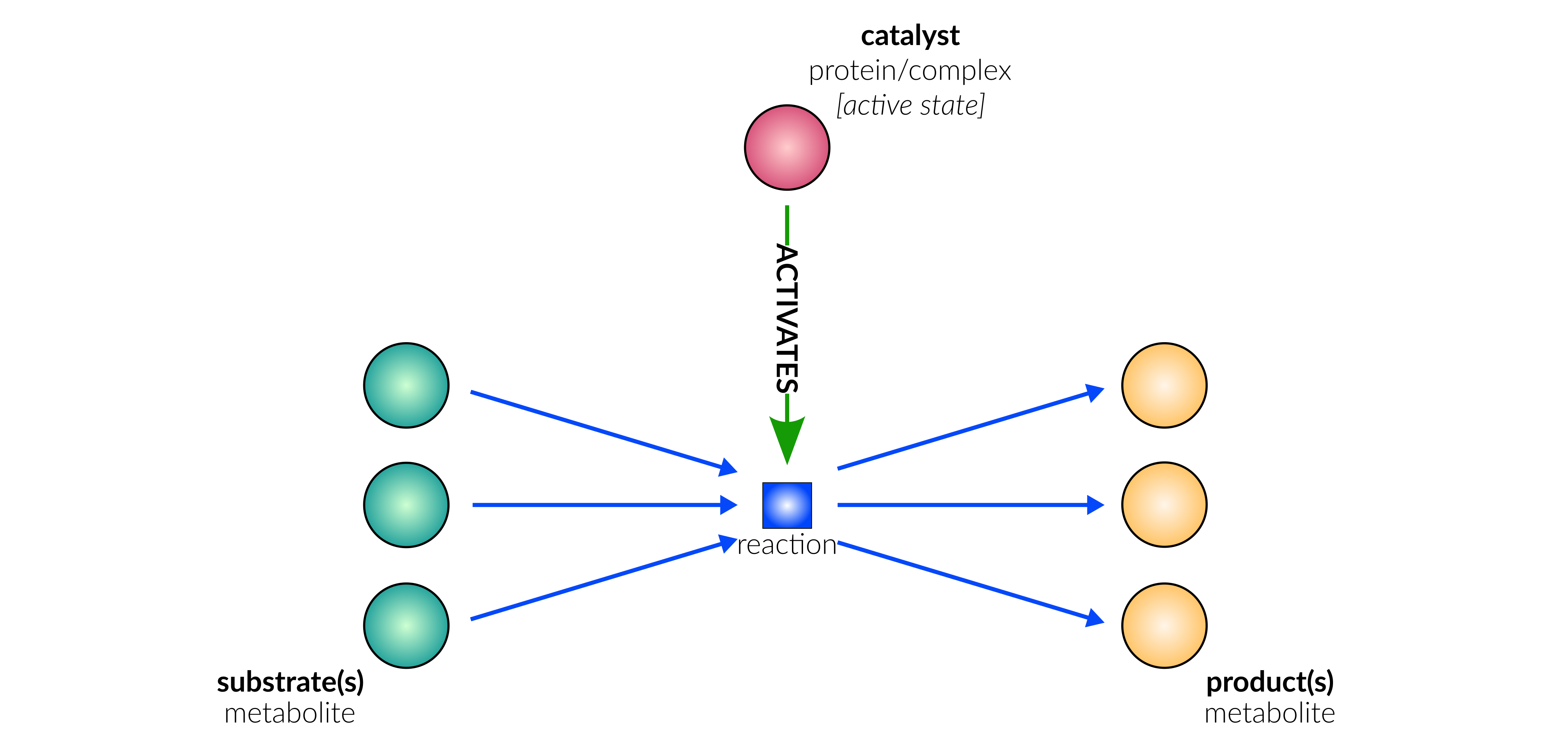
degradation/secretion
Biological representation
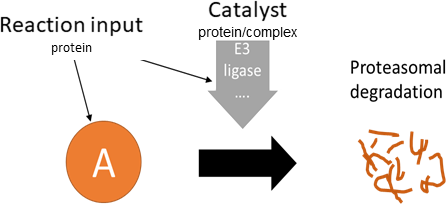
Database representation
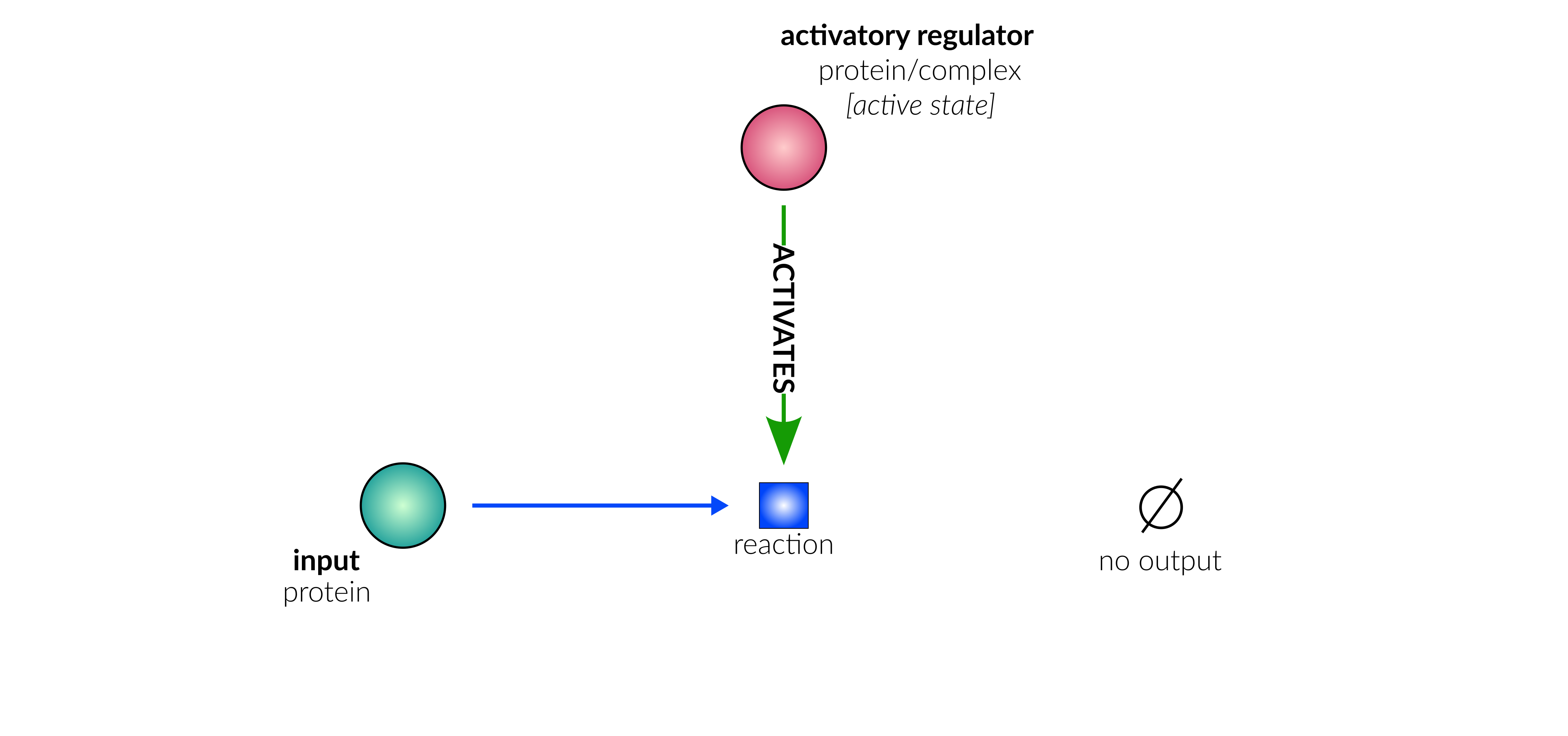
protein deactivation
Biological representation
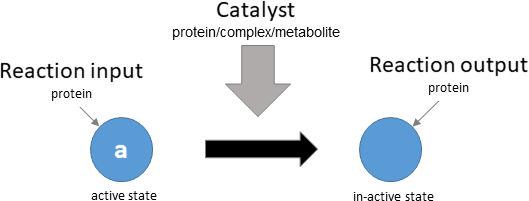
Database representation
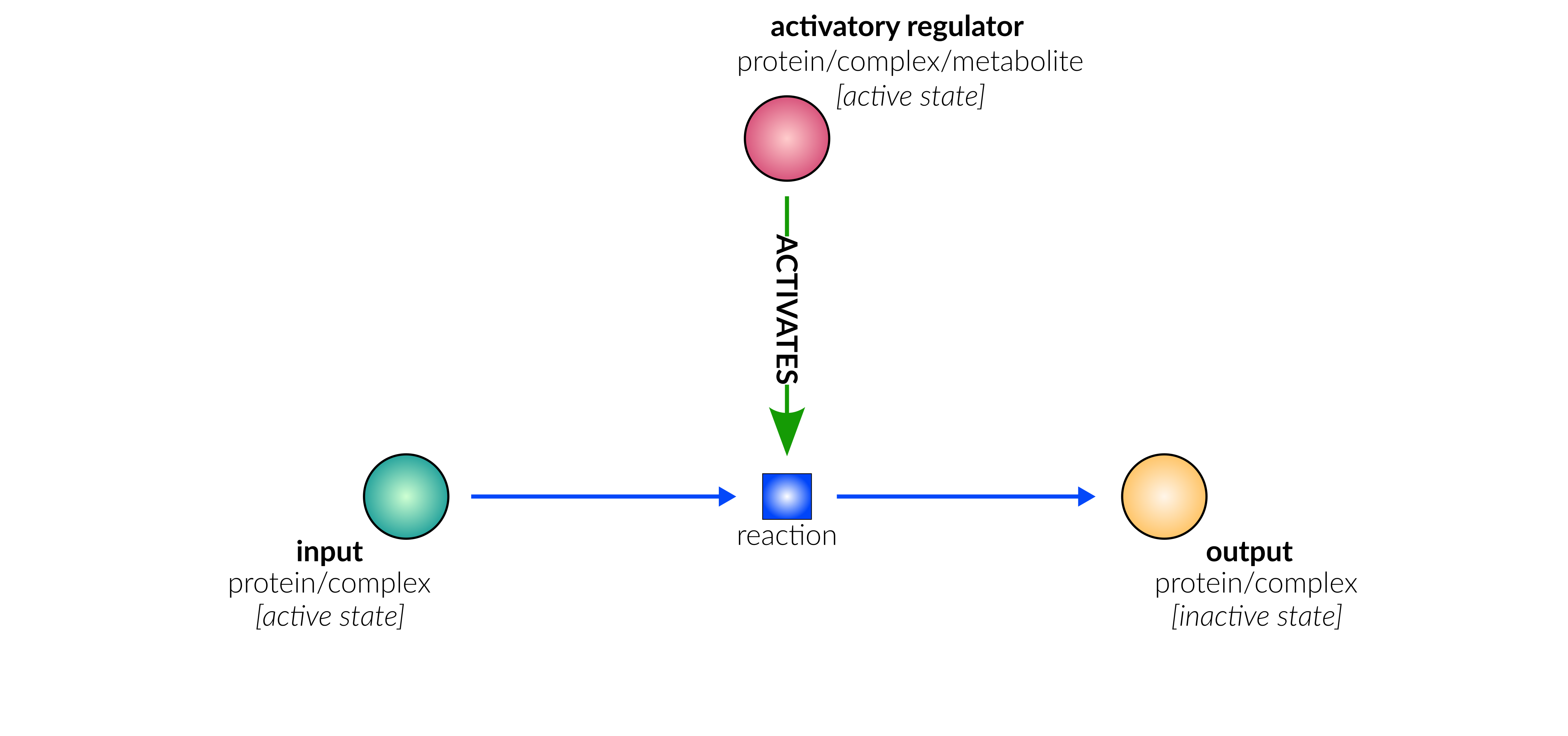
protein activation
Biological representation
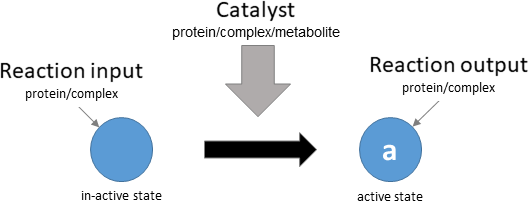
Database representation
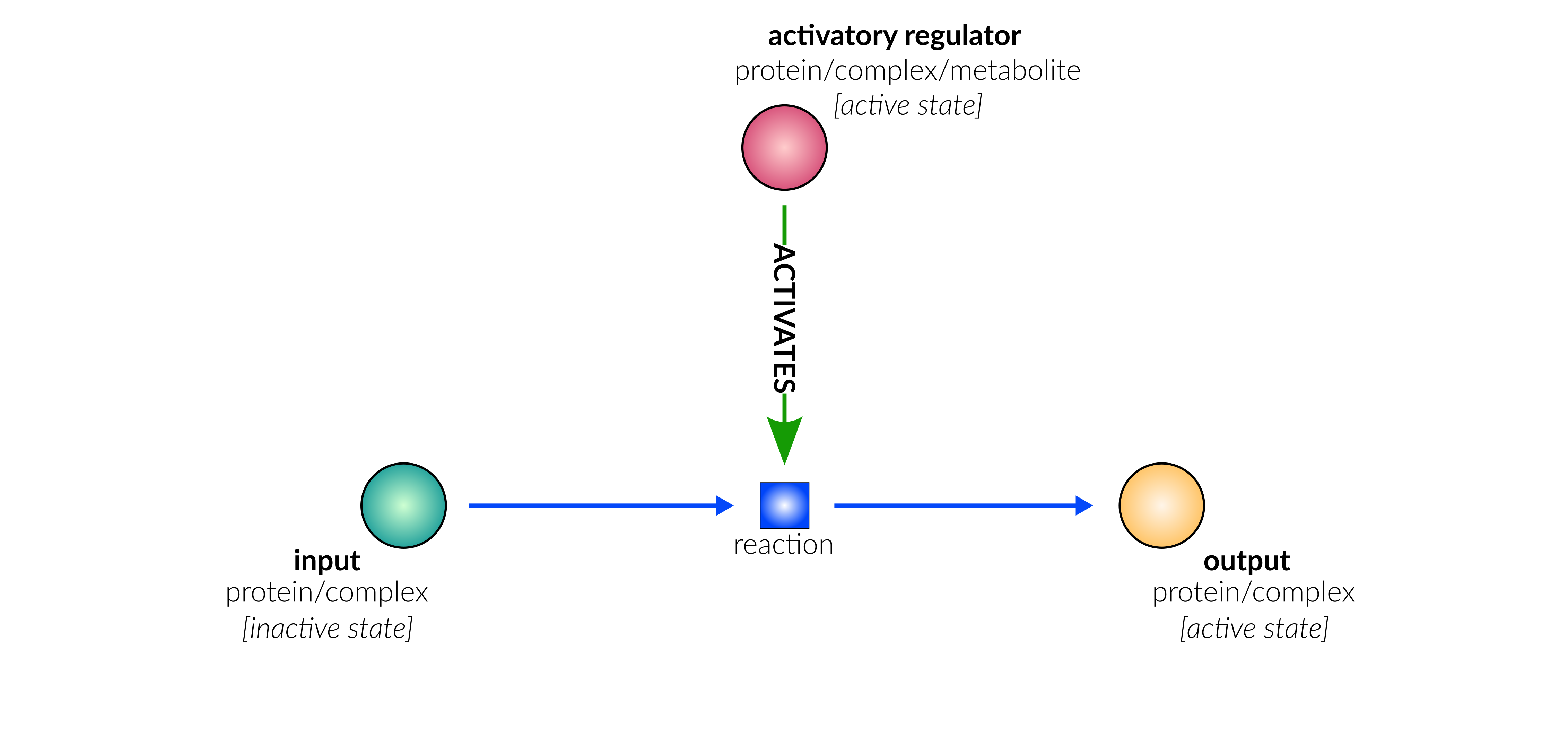
transcriptional/translational activation
Biological representation
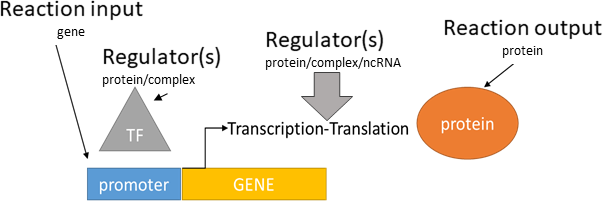
Database representation
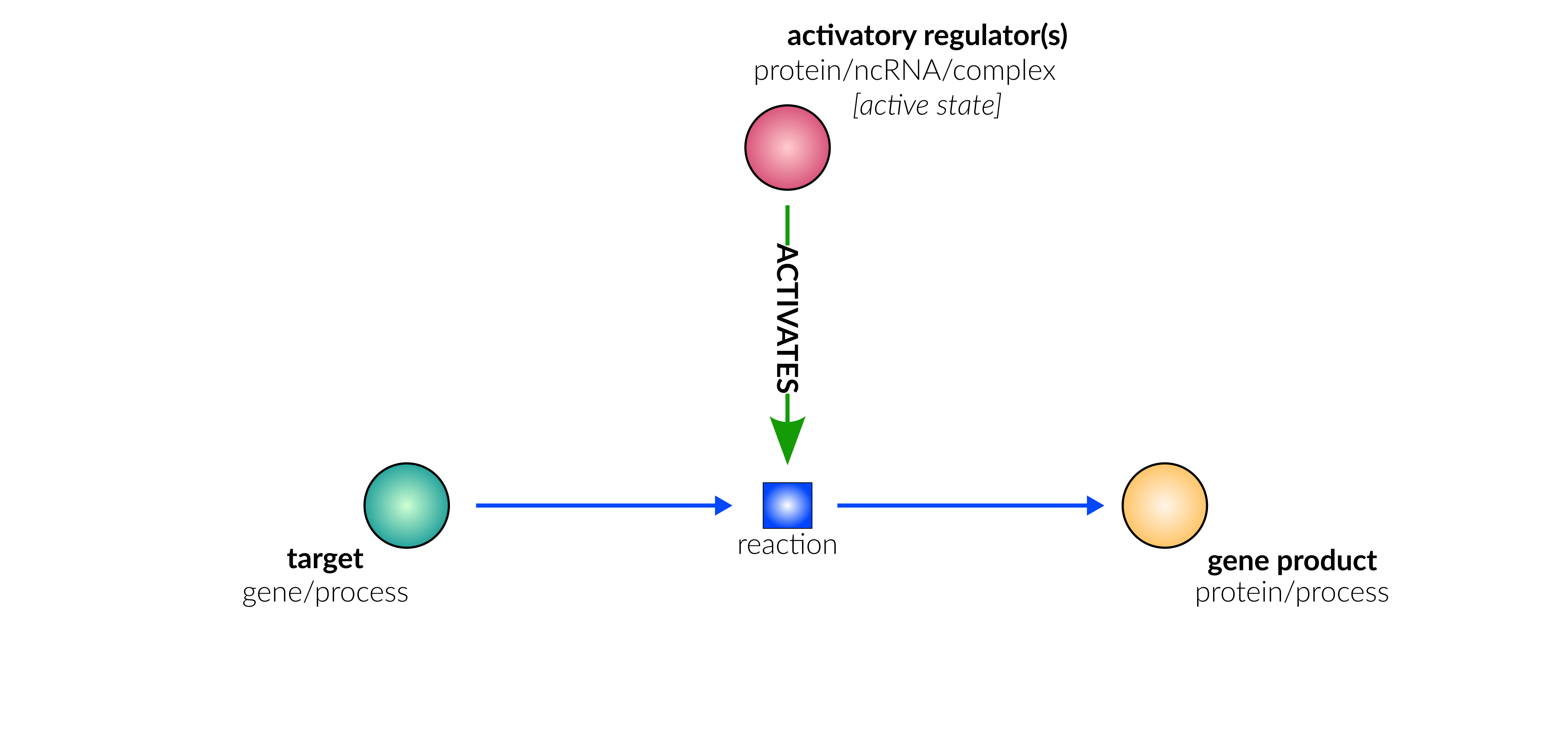
transcriptional/translational repression
Biological representation
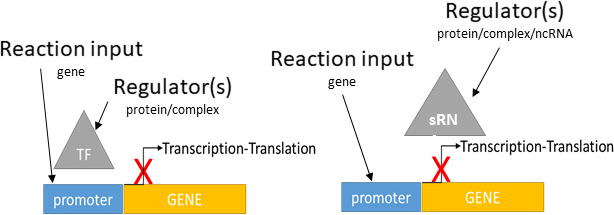
Database representation
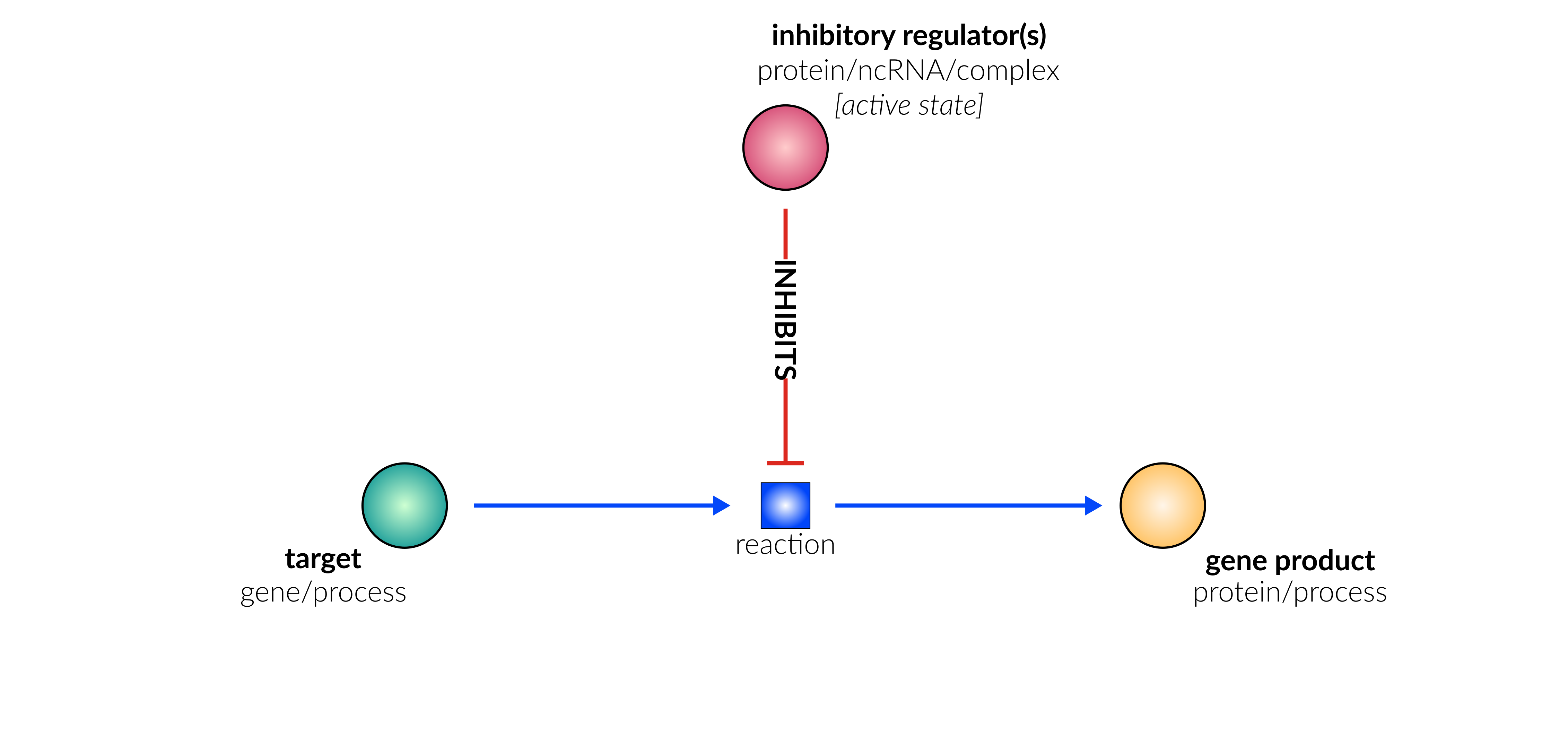
translocation
Biological representation
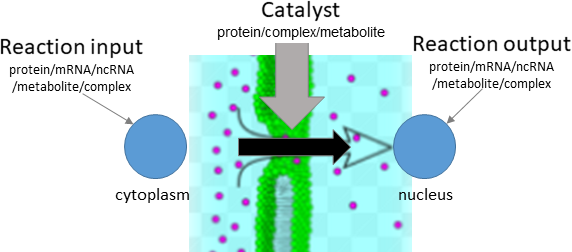
Database representation
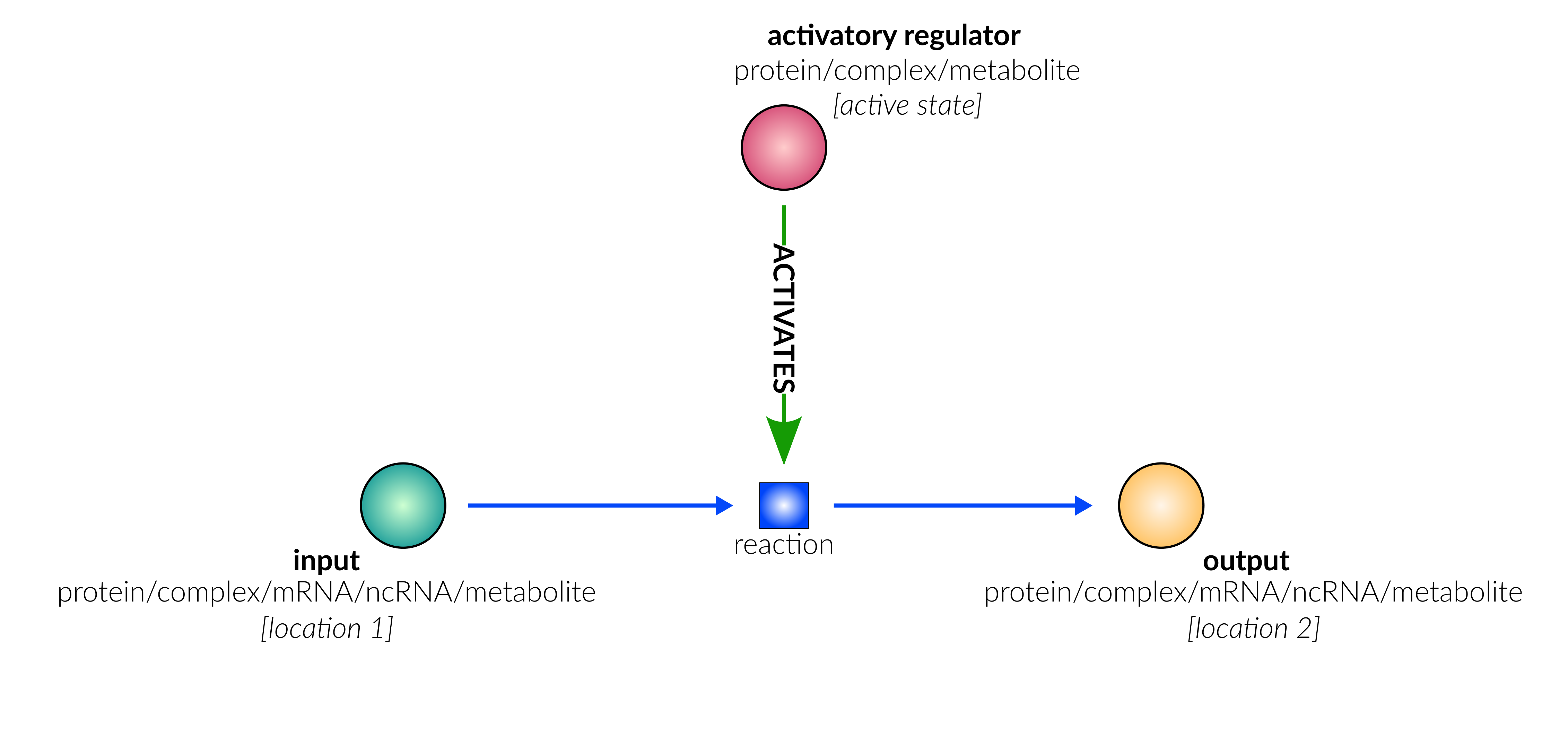
unknown
Biological representation
Database representation